|
Biomolecules Folding and Disease |
|
|
The course Laboratory of Bioinformatics 1 (66563) includes in the second semester a 60-hours module given by
Emidio Capriotti.
The course director is Prof. Rita Casadio.
The module is running from March 13 to May 4, 2017 in the Laboratory of Bioinformatics at the ground floor of Via San Giacomo 9/2 (back entrance).
This page will include all the slides and materials of the lectures.
- Materials
Course Schedule Introduction and Basic Concepts March 13, 2017. Protein Structure Alignment March 14 and 16, 2017. Multiple Alignments March 20 and 21, 2017. Hidden Markov Models March 23, 2017. HMM Alignments March 27, 2017. HMMER March 28 and 30, 2017. Introduction to the Project April 3,4 and 6 2017. Project Implementation April 10,11,20 and 21 2017. Protein Structure Prediction April 27 and 28 2017. Structure Analysis and Variants May 2 2017. Predicting the Effect of Protein Variants May 4 2017. - Requirements
- References
- Evaluation
| This is a theoric and hands-on module which requires the use of PC. Students need to use their own laptop. Practical session of the course will be performed on a Linux OS. |
|
Structural Bioinformatics II Edition:
Structural Comparison and Alignment. by Marti-Renom et al. |
|
A Comprehensive Benchmark Study of Multiple Sequence Alignment Methods: Current Challenges and
Future Perspectives. PLOS ONE, 2011. by Thompson et al. |
|
Pfam: the protein families database.
Nucleic Acids Research, 2014. by Finn et al. |
|
Assignment of protein sequences to existing domain and family classification systems: Pfam and the PDB.
Bioinformatics, 2012. by Xu and Dunbrack. |
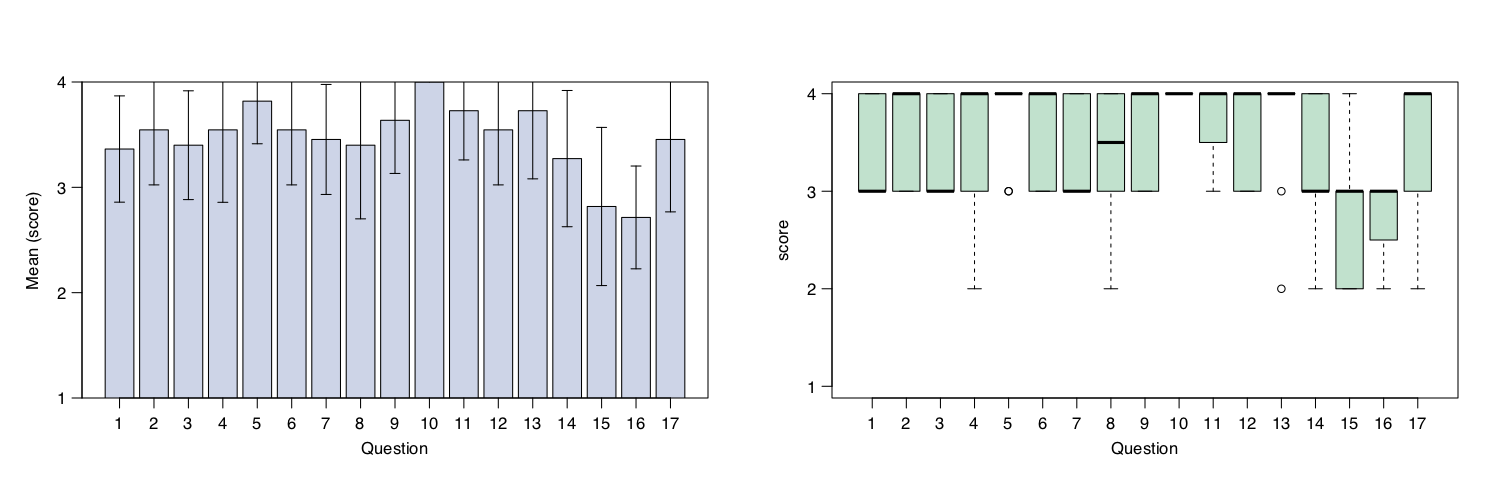
In this page we included the
evaluation form anonymously compiled by the
participants to evaluate the quality of our course. Answers submitted with
numbers from 1 (No) to 4 (Yes) have been included in the following statistics
calculated by R. The evaluation form was compiled by 11 participants which rated
the course with an average score of 3.5 over 4.
The received positive and negative comments will be published soon.
 Data |
